New and updated fragment databases from ChEMBL and SureChEMBL are now available in Spark.
This new release of databases significantly expands the available
chemistry to Spark users by including the latest compounds from
scientific literature, as well as previously unseen compounds from
chemical patents. Combined with databases of fragments derived from screening compounds, and with custom databases which you can generate from your corporate collections using the Spark Database Generator, they provide an outstanding source of bioisosteres you can use to generate new ideas for your project.
More than 1.6 million fragments from ChEMBL
The updated Spark ‘ChEMBL’ databases include an expanded choice of more than 1.6 million fragments derived from release 30 of ChEMBL, a collection of around 2.1 million compounds reported in peer-reviewed scientific literature.
More than 14 million fragments derived from SureChEMBL
In addition, new ‘SureChEMBL’ fragment databases are available to
Spark users for the first time. These include more than 14 million
fragments derived from the SureChEMBL
collection of approximately 17 million compounds harvested from the
patent literature. This addition affords Spark users a significant
expansion on fragment databases derived from ChEMBL, providing a broader
range of accessible chemistries than ever before.
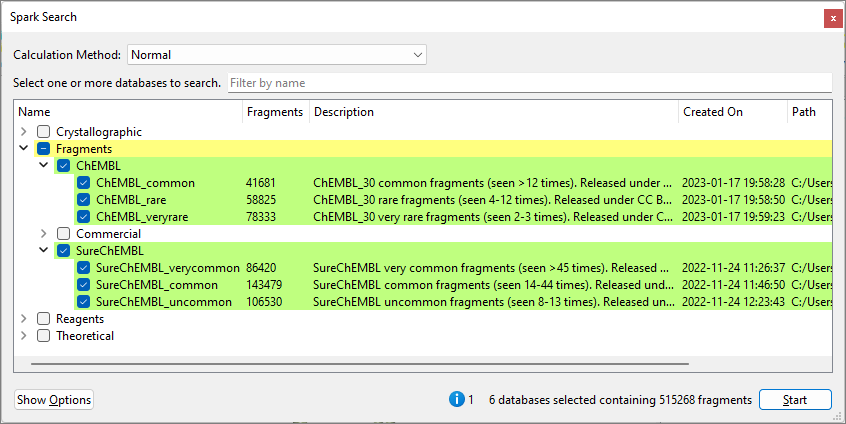
Figure 1. Spark Search window for an
R-group replacement experiment, showing default ChEMBL and SureChEMBL
fragment collections with a single attachment point.
Like ChEMBL, SureChEMBL compounds come with property information such
as synthetic tractability, toxicity, and metabolic reactivity.
Compounds in the original source collections were filtered on these
biochemical properties prior to fragmentation in Spark, removing
molecules containing potentially toxic or reactive groups. Compounds
were then cleaved by breaking the bonds connecting carbon atoms to
functional groups such as heteroatoms, carbonyls, thiocarbonyls and
rings, whilst preserving functional groups such as carboxylic acids,
nitro groups and rings. All fragments were subject to heavy atom count
and rotatable bond limits, which increases the probability that
fragments will form part of biochemically active small molecules.
Finally, fragments were sorted by frequency of occurrence in the source
dataset and grouped by commonality to form Spark databases. A key
assumption for Spark users is that regularly occurring fragments are
likely to form part of novel active biochemistry, therefore we recommend
that users try the more common libraries before moving on to less
common.
Table 1 reports the total number of fragments in each Spark database,
with the frequency of occurrence in the source dataset. Database
frequencies were arbitrarily chosen to give manageable database file
sizes.
Table 1. Fragment databases sorted by frequency.
| Spark category | Database | Total number of fragments (x1000) | Frequency in source dataset |
| ChEMBL | Common | 223 | Fragments which appear in more than 12 molecules |
| Rare | 304 | Fragments which appear in 4-12 molecules | |
| Very Rare | 390 | Fragments which appear in 2-3 molecules | |
| Extremely Rare* | 783 | Fragments which appear in 1 molecule | |
| SureChEMBL | Very Common | 509 | Fragments which appear at least 45 molecules |
| Common | 795 | Fragments which appear in 14-44 molecules | |
| Uncommon | 554 | Fragments which appear in 8-13 molecules | |
| Rare* | 957 | Fragments which appear in 5-7 | |
| Very Rare* | 757 | Fragments which appear in 4 molecules | |
| Extremely rare* | 979 | Fragments which appear in 3 molecules | |
| Doubleton* | 2,545 | Fragments which appear in 2 molecules | |
| Singleton* | 4,794 | Fragments which appear in 1 molecule |
*Contact us for further details.
The number of compounds per connection point count in each database is presented in Figure 2.
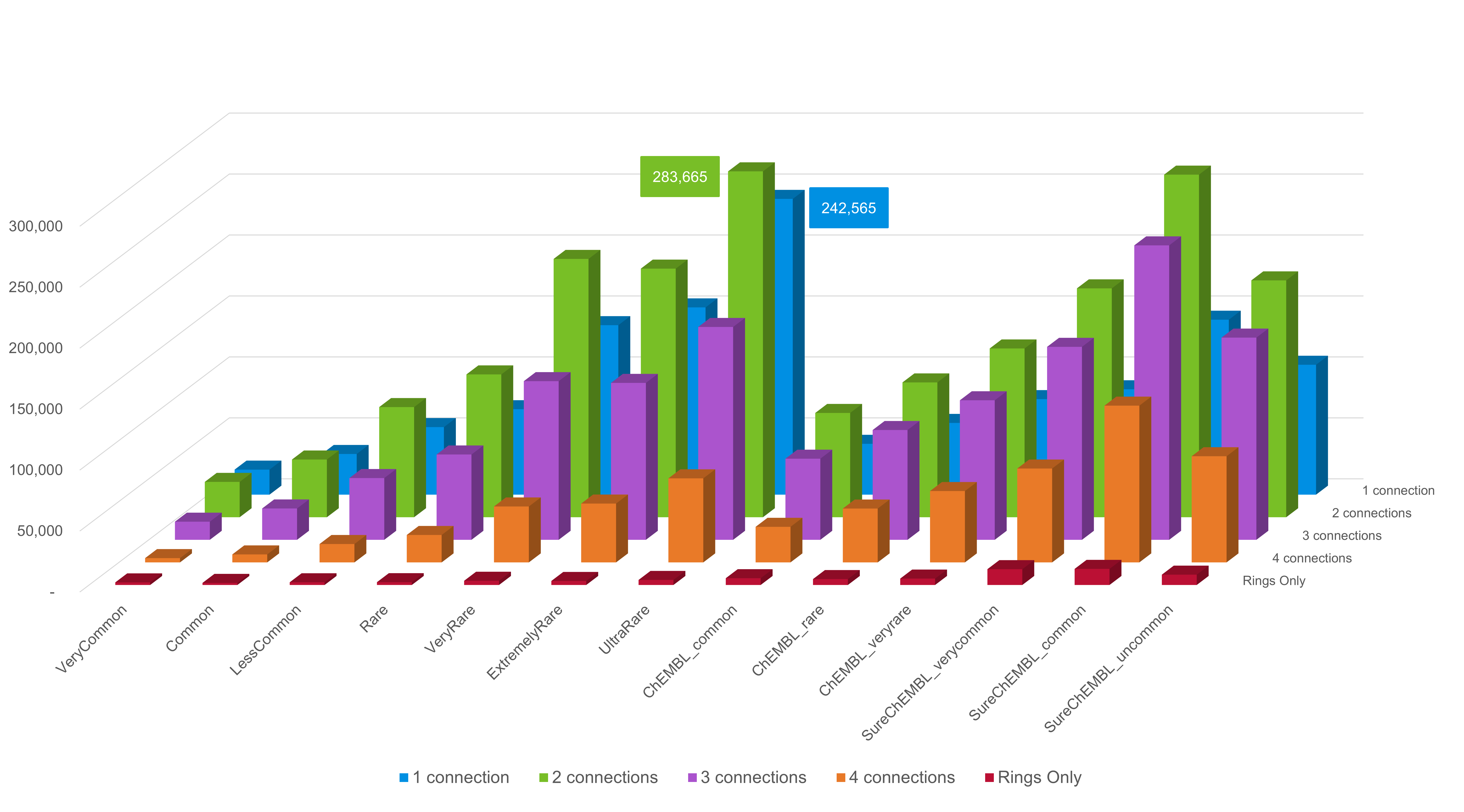
Figure 2. Count of fragments in the
recommended Spark Commercial, ChEMBL and SureChEMBL databases split by
the number of connection points on each fragment.
We recommend that users only install ChEMBL databases where the
fragment frequency is at least 2, and the three SureChEMBL ‘Common’
databases (Very Common, Common and Uncommon), as the size of the
SureChEMBL databases is very large. Furthermore, singleton and doubleton
databases for both ChEMBL and SureChEMBL may contain fragments derived
from erroneous structures in the source dataset.
Though ChEMBL and SureChEMBL fragments are from different sources
(scientific literature vs. patents), as expected, there is significant
overlap between some of the databases, as shown in Table 2. In
particular, the ChEMBL Common and SureChEMBL Very Common databases show
the highest overlap, which is to be expected because most bioactive
compounds contain basic subunits, e.g., phenol or pyridine rings.
Table 2. Overlap of the most common fragments in the ChEMBL and SureChEMBL databases.
| SureChEMBL | ||||||||||
| Very Common | Common | Un-common | Rare | Very Rare | Extremely Rare | Doubleton | Singleton | Unique | ||
| ChEMBL | Common | 66% | 14% | 3% | 3% | 1% | 1% | 2% | 2% | 8% |
| Rare | 28% | 21% | 7% | 7% | 3% | 3% | 4% | 5% | 23% | |
| Very Rare | 13% | 17% | 7% | 8% | 4% | 5% | 7% | 6% | 33% | |
| Extremely Rare | 6% | 10% | 6% | 7% | 4% | 5% | 8% | 13% | 42% | |
Reagent databases
The January update of the Spark reagent databases includes around 293,000 reagents derived from the eMolecules building blocks using an enhanced set of rules for
chemical transformation. These databases are updated monthly to give
you up-to-date availability information, to make it easy for you to
order the reagents you require to synthesize your favorite Spark
results.
Update your Spark databases
The updated ChEMBL and new SureChEMBL databases, combined with the Spark Commercial fragment databases,
provide more than 19 million unique fragments to search, with more than
4.6 million fragments in the recommended databases alone. They
significantly expand the choice of fragments for your Spark experiments,
providing an even better source of novel ideals for your drug discovery
projects.
Spark users can contact our Support team to update their ChEMBL and SureChEMBL databases.
If you are a medicinal chemist or computational chemist and not currently using Spark, contact us to find out how it can help you generate innovative ideas, explore chemical space and escape IP and toxicity traps, or request an evaluation to try Spark on your project.