The field of protein DNA/RNA interactions is an expanding area of research. These interactions affect fundamental processes and as a result, relevant protein DNA/RNA processing complexes and their associated mechanisms provide interesting therapeutic targets to researchers. However, due to the complex and often delicate nature of these systems, traditionally researchers haven’t had sufficient tools to address them.
Cresset Discovery has a proven track record in delivering projects around DNA/RNA interacting proteins. Our team of expert modelers has access to the specialist computational approaches and knowledge to address these challenges and leverage the best possible outcome for your project.
The importance of protein DNA/RNA complexes
Selectively targeting DNA or RNA has proved challenging and the effect of DNA-binding drugs on cells is analogous to a shotgun blast with multiple off-target effects which are difficult to predict. Therefore, the focus of drug discovery firstly moved towards protein-protein interactions which brought its own pitfalls and problems. Now the focus is in interacting with the protein DNA/RNA complex and the mechanism of protein processing of DNA/RNA. This has been aided by the increasing amount of structural data available especially from the rapidly expanding field of Cryo EM.
Protein DNA/RNA complexes are important for many areas of drug discovery and are active against multiple disease types. For example, they are the target for the fluoroquinolone class of antibiotics (as shown in Figure 1) which display the intercalation mode of ligand binding were the ligand sits between adjacent DNA/RNA base pairs.
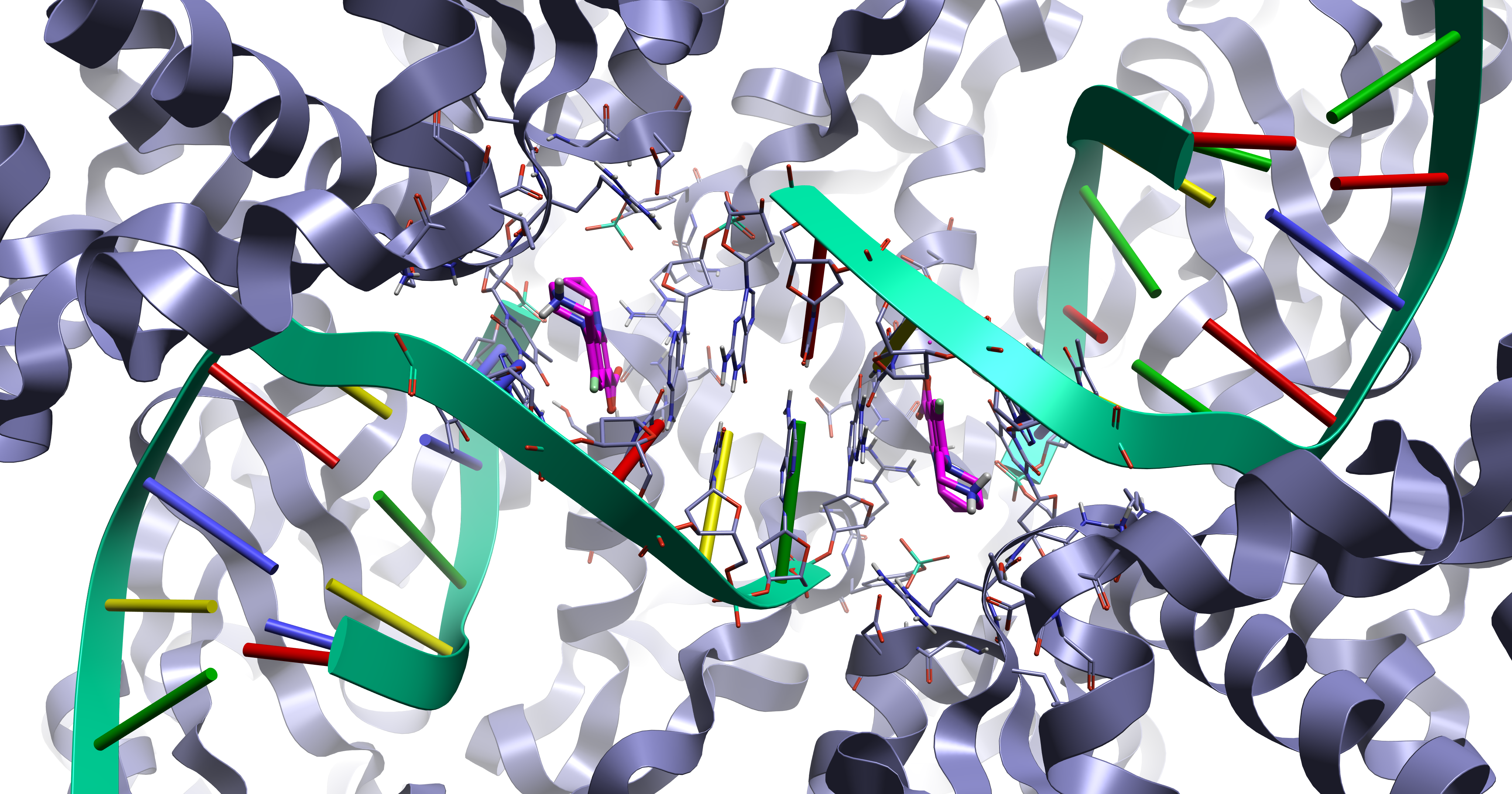
Figure 1. The 5btc1 crystal structure displaying the fluoroquinolone intercalation DNA binding mode.
Understanding protein DNA/RNA mechanisms
How proteins process DNA/RNA varies both across and within protein families. The proteins that process DNA/RNA are components of large molecular ensembles which split, cleave, and process DNA/RNA. Whilst it is not always possible to understand the full processing mechanism and all the individual components involved, it is important to have a least the basic outline of the mechanism of DNA processing and principal of the synergistic DNA/RNA and protein movement involved. At Cresset Discovery, we have experience of identifying and handling many of these different processing mechanisms whilst identifying hotspot features which can be used to disrupt the mechanism.
A particularly interesting example of DNA/RNA interacting proteins are helicases which are a large family of proteins thought to be critical to all organisms for the processing of DNA and RNA. Helicases move along the DNA or RNA strand and using an ATP-driven process, split the double strand into two single strands. There are 6 superfamilies of helicases described using their sequence motifs, functionality, and mechanism of action. Helicases can adopt multiple different conformations and oligomerization states with which to process DNA or RNA which will be dependent on the specific helicase under investigation. Multiple different processing mechanisms have been identified for how different helicase process DNA or RNA and understand which mechanism and the important conformational state is a key factor in a successful project.
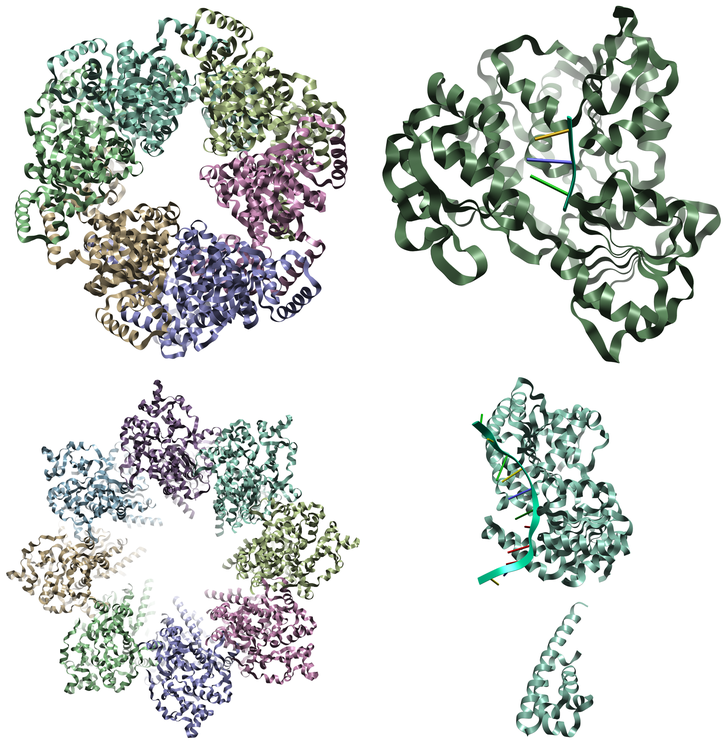
Figure 2. Examples of different states of different helicase structures: Bloom syndrome protein (upper left), Vibrio cholerae replicative helicase (upper right), Octameric Human Twinkle helicase (lower left) and ZIKV NS3 helicase (lower right).
Helicases are just one example of DNA/RNA interacting proteins, which also include amongst others High Mobility Group Proteins (HMGP), DNA Methyltransferases, Polymerases, Topoisomerases, and Transcription factors which all exhibit distinct and interesting methods of interacting and processing DNA. These proteins represent potentially very interesting drug targets against a wide range of diseases.
Computational processing of DNA/RNA – protein complexes
At Cresset Discovery, we have gained experience across numerous projects in working with DNA/RNA – protein complexes and we understand how delicate these types of systems can be if not treated with care and respect. The highly charged nature of the DNA/RNA phosphate backbone tends to be matched with corresponding lysine and arginine residues from the protein along the interaction face, and correctly assigning the interaction profile is the first challenge. Mixed in with these charge-charge interactions are more subtle interactions which further help to stabilize the complex. The addition of counter metal ions can also be required to stabilize the DNA/RNA backbone that are close to the protein as well as regions that are not fully coordinated to the protein. The inclusion of key water interactions further helps to improve the quality of the model. Careful processing of the complex enables Cresset Discovery modelers to employ the use of computationally more intensive methods like Flare™ Molecular Dynamics to assess complex structure reliability.
When undertaking computational ligand selection, automated methods tend to favor highly charged system which are non-druglike, due to the phosphate backbone and complementary protein residues which bias computational methods toward corresponding functionalities bases and acids respectively. Here, our team applies the medicinal chemistry insight needed in the post-processing of results to identify suitable compounds which are lead like, non-highly charged and suitable for progression.
Engage Cresset Discovery to accelerate your project
Cresset Discovery serves as a true partner for computational modeling in drug discovery. Our expert team has a breadth of experience across medicinal and computational chemistry, and biology to work alongside you to deliver your project goals.
To learn more about how we can support you with protein DNA/RNA interactions or any other project, contact us or request a no-risk free confidential discussion with our team.
References
- Blower, T.R., Williamson, B.H., Kerns, R.J., Berger, J.M.; Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis, PNAS. 2016, 113 (7), 1706-1713.