Free Energy Perturbation (FEP) methods are a fast-evolving area of drug discovery, and Cresset Discovery’s dedicated team of expert modelers are establishing a rapidly growing portfolio of successfully completed FEP projects. There are a number of key data requirements that are necessary for an FEP project to have the best chance of success, and in this article, we outline the ideal starting point for running an FEP calculation. However, where this level of data is not available, our team can work with you to assess whether FEP, or other computational techniques are more appropriate. With our deep knowledge of free energy theory and method implementation, Cresset Discovery will identify the best approach to accelerate your project.
The three ideal data requirements for an effective FEP calculation can be characterized as:
- The availability of a high-quality crystal structure with a relevant ligand
- The quantity and quality of a dataset of ligands for validation
- The generation of an appropriate ligand set for prediction
Here, we describe these requirements and best practices for FEP calculations in detail.
Availability of a high-quality crystal structure
To effectively implement FEP, it is ideal to have access to a high-quality crystal structure with a known ligand, relevant to the ligands in your data set, bound along with extended knowledge of the binding mode interactions. What is crucial, in all relative FEP calculations, is that the binding mode of the ligand is unambiguously defined within the protein structure. While a crystal structure to a resolution below 2.2 Å is ideal, ligand occupancy, unresolved sidechains, and overall quality of the structure must also be evaluated.
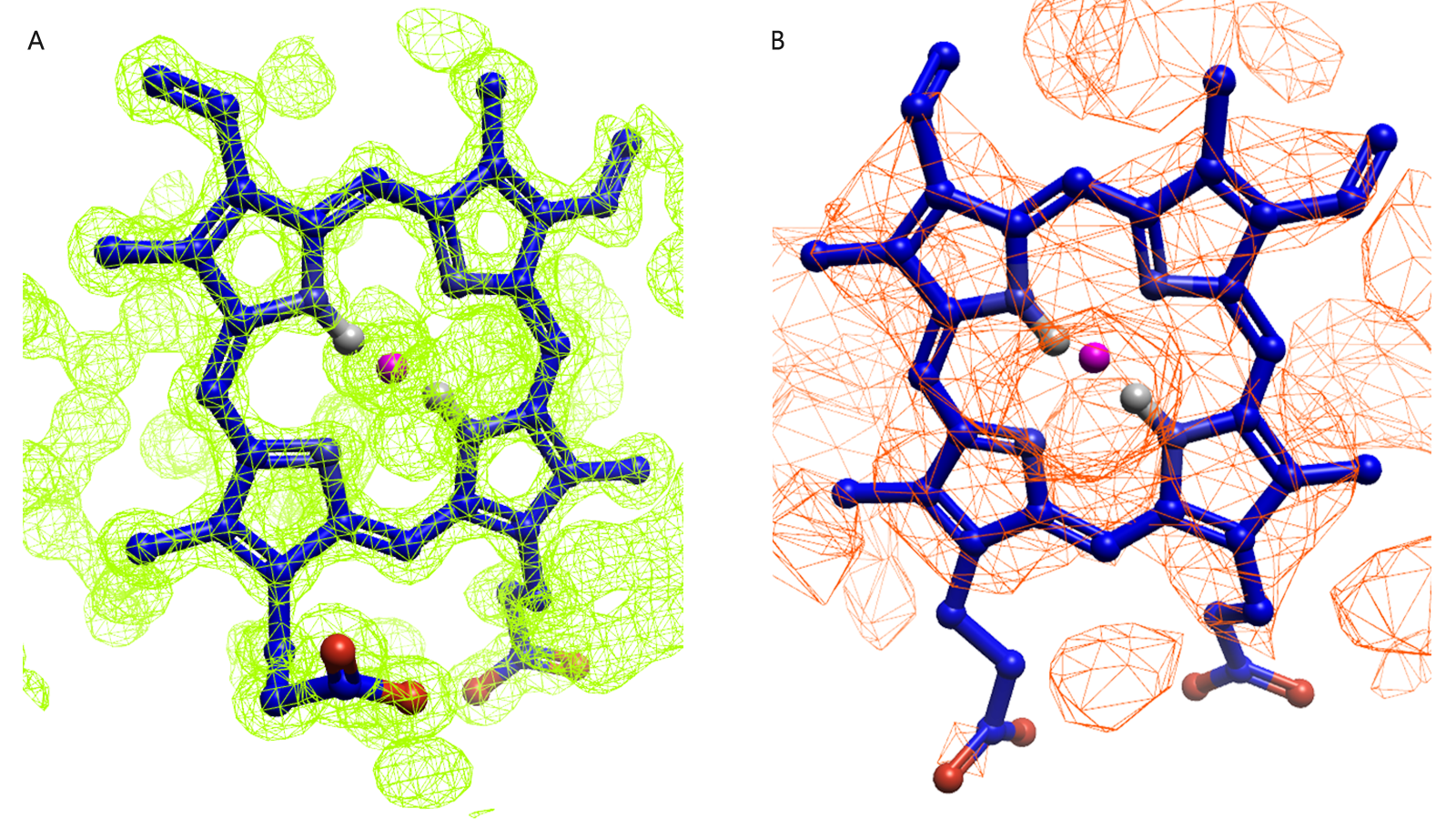
Figure 1. Example of protein structures with good (green surface) and poor (red surface) electron density maps for the same target. (A) Protoporphyrin ligand from myoglobin, 1A6M PDB ID (1.0 Å resolution). (B) Protoporphyrin ligand from hemoglobin, 1S0H PDB ID (3.0 Å resolution). Image generated with Flare™ v7.
Like all structure-based techniques, the binding mode plays a crucial role in FEP calculations when predicting the activity of a set of ligands. The binding mode of the known ligand serves as a reference, to which the novel ligand binding modes and poses are compared.
Examining electron density to observe a well resolved and known binding mode and pose can be helpful in choosing input for your FEP calculation. The example provided in Figure 1 serves as an illustration of this analysis and how, in such an example, choosing PDB 1A6M (Figure 1A) is likely the better choice in terms of picking a ligand binding event you have confidence in.
Adequacy of ligand data for validation
FEP, as with other computational methods, requires validation, and this relies on a dataset of experimentally measured ligand data for the system under investigation. This validation stage, otherwise known as ‘benchmark’ mode, is an initial test of FEP to determine how well the predictions match experimental data. If there is a good match, this gives confidence in the FEP set up and model, such that it is worthwhile moving into the ‘production’ mode of making predictions based on no experimental measurements. Appropriate ligand data used in benchmarking are typically derived from the following types of experimental data:
- XC50 (half-maximal activity concentration),
- Ki (inhibition constant), or
- Kd (dissociation constant).
Critical evaluation of the diversity and reliability of the ligand dataset is central to the validation of the FEP calculation i.e., the quality of the benchmark. It is preferable to have a ligand set with a wide range of binding affinities, including both weaker and stronger binders, to capture a broader spectrum of ligand-protein interactions, and to validate the FEP predictions fully. An accuracy of < ± 1 kcal/mol of predicted affinity to the experiment can be achieved in FEP, and this will be established in the benchmark validation. Note, in the analysis of the benchmark study, it is worth taking into account how reliable the experimental data is, and again ideally, where possible it is preferable to use single source assay information to avoid large uncertainty in the experimental results that can arise due to variation in conditions.
Presence of appropriate ligands for prediction
Based largely on the results of the benchmark study the FEP project can move into a production mode: the model is ready to make predictions based on new molecular designs. As in the benchmarking, the diversity and suitability of the ligands for FEP calculations must be evaluated for the production calculation. Testing a set of new ligands which are very structurally dissimilar will not work well in relative FEP given the assumption stated above.
The structural similarities between the reference ligand and the predicted ligands must be assessed to establish whether they share key structural features, common substructures, and consistent binding mode. Suitable similarity ensures that the FEP calculations provide meaningful and reliable predictions for the ligands of interest. An illustration of such an analysis is provided in Figure 2.
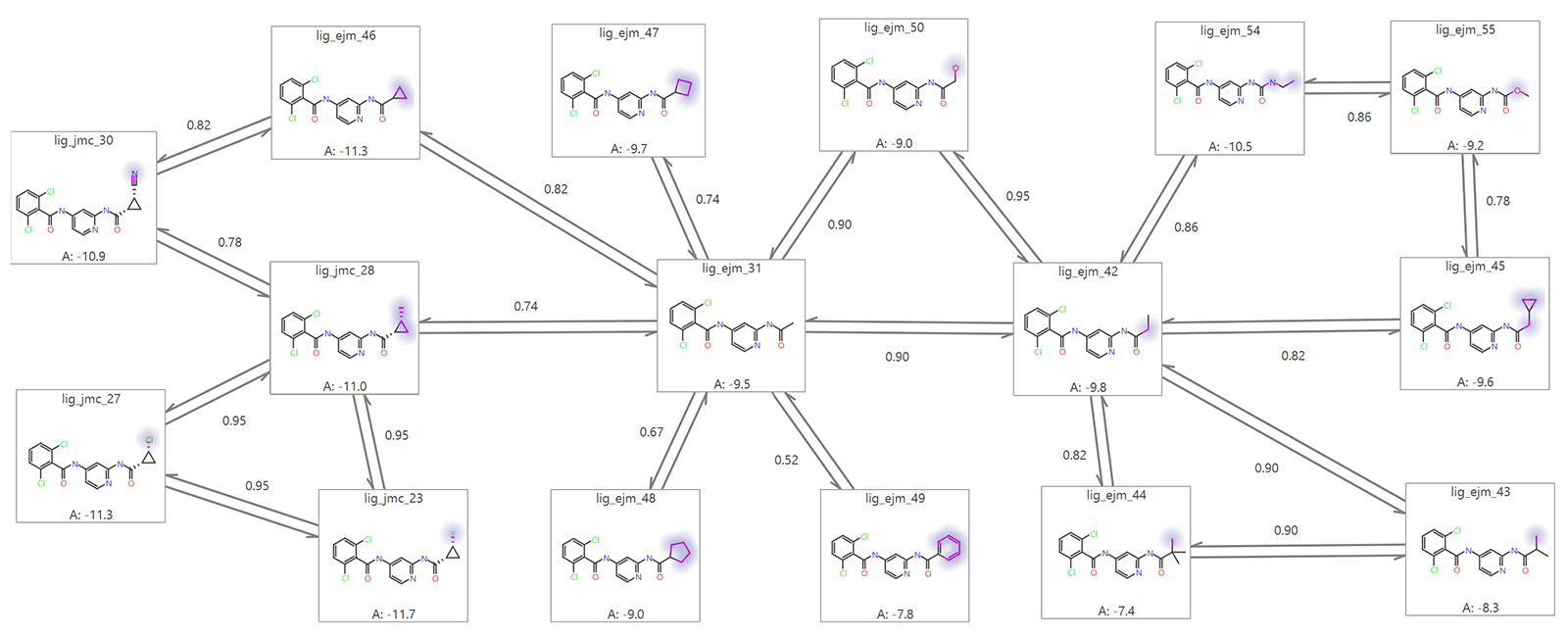
Figure 2. Examples of permitted alchemical changes within an FEP calculation. Image generated with Flare.
Figure 2 illustrates an FEP graph which links up ligands by various transformations. Each link has a ‘score’ which ranges from 0 (no similarity) to 1 (identical) and so gives an indication of the ease of the perturbation; a higher link score (typically > 0.4) indicates a link (transformation) more likely to calculate successfully. In terms of setting up an FEP calculation for production mode and making successful predictions for a new ligand data set, it is important to check you have a network of links with good (> 0.4) link scores. Very low link scores represent a data set with too much structural diversity. Cresset Discovery scientists have the expertise and experience to guide you in creating such a dataset.
Determining if FEP is suitable for your project
The feasibility of implementing FEP calculations for your project depends on several key factors. The availability of a high-quality crystal structure with a relevant ligand, a well-defined binding mode, and adequate ligand data for validation can be critical elements for accurate and reliable FEP calculations. However, success with FEP can still be achieved even in the absence of certain information.
The presence of appropriate ligands for prediction, with structural similarities to the reference ligand and a range of binding affinities, enhances the validity and applicability of FEP predictions. When these ligand requirements are met the amenability of your target to FEP generally can be initially assessed with a benchmark study. Careful evaluation and consideration of the benchmark run will guide the decision on whether FEP can be effectively utilized to support your project’s objectives, in a production run (or predictive mode) or if alternative approaches should be explored.
Alternative methods, such as ligand-based approaches, bioisosteric replacement, and de novo design, can be considered in cases where the requirements for FEP are not fully met. Expertise at Cresset can help you evaluate the suitability of your project for FEP.
Successful application of FEP using Cresset methodology is demonstrated in a benchmarking study which accurately calculated binding affinities for a dataset of 30 ligands that bind between the lipid and GPCR interface in P2Y1. In this study, predicted binding affinities agree with experimental measurements and are in line, or better than data published in the literature. On the basis of this, the FEP model can be taken forward with confidence and used to test and predict new designs.
Cresset Discovery’s experienced team is committed to providing the most appropriate method for developing your project. Contact us for a confidential discussion about your goals and we can advise on whether FEP would be suitable for your project or if other, more appropriate computational methods would be suitable to progress your project.